Model building
This section covers model options configuration, model building execution and model file information.
Model options
On “Model options” window, you can configure the model parameters to make a better model. There are 4 tabs – “Normal”, “Binary model”, “Regression model”, “Multiclassification model” on which you configure options to build different types of models.
Normal
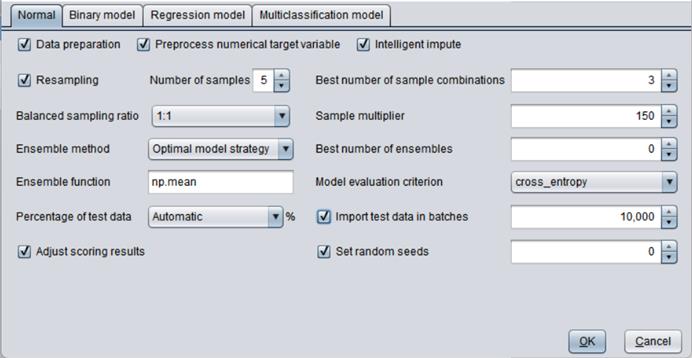
“Data preprocessing”: Preprocess data or not before performing data modeling;
“Preprocess numerical target variable”: You can choose whether preprocess a numerical target variable before performing data modeling or not.
“Intelligent impute”: Intelligently assign a value to a missing value.
“Ensemble method”: “Optimal model strategy” and “Simple model combination”. The former selects best top N models to build a new model and involves relatively a large amount of computations. The latter just combines all defined models to build a new model and involves a relatively small amounts of computations.
“Best number of ensembles”: Select the best model combination. 0 means selecting the most effective model among the combinations; >0 means selecting the fixed top N models; and <0 means selecting the one among the top N models that makes the most effective combination. Default is 0.
“Ensemble function”: The option specifies the approach of combining models. You can select any function included in numpy. Default is np.mean.
“Model evaluation criterion”: Use AUC, cross_entropy, KS, AP, Recall, Lift, f1_score for a binary model and default is AUC; use MSE, MAE, MAPE and R2 for a regression model; and use cross_entropy for a multiclassification model.
“Percentage of test data”: Test data percentage.
“Import test data in batches”: Set the number of rows of test data to be imported in each batch.
“Adjust scoring results”: By default, the model scoring results will be adjusted according to the average of the sample data. Without adjustment the score is the average of the balanced samples.
“Set random seeds”: You can control randomness of model building through this option; default is 0. If the value is null, two model building executions will get random results respectively. If the value is set as integer n, two executions will get same results if their ns are same; and different results if their ns are different. When the random seed is set as n, the random_state value of all models will be set as the same value and can’t be manually changed.
Binary model
You can configure parameters for binary models on “Binary model” tab. A selected binary model will be used for model building.
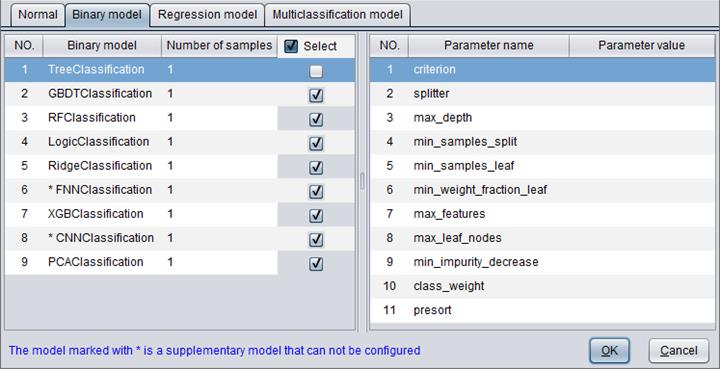
There are 9 types of binary model: TreeClassification, GBDTClassification, RFClassification, LogicClassification, RidgeClassification, FNNClassification, XGBClassification, CNNClassification and PCAClassification.
“Number of samples” determines the number of samples used to build a model.
Below are parameter configuration directions for binary models.
Appendix 1: Binary model parameters
Type & Range: The type is always followed by an interval indicating the parameter’s value range. Square brackets indicate a closed interval and parentheses indicate an opened interval. Braces are used for certain int and float parameters to represent drop-down-menu values; the format is {start value, end value, interval}, such as {1, 5, 1}=[1,2,3,4,5] and {1, 6, 2}=[1,3,5]. All available values of string parameters will be listed in the drop-down menu and can’t be entered manually; a null value should be selected through the drop-down menu; the bool values, which are true and false, also should be selected in the drop-down menu; values of int parameters and float parameters can be enterned manually and all available values will be listed in drop-down menu.
For some float parameters, if their values are integers, they need to be followed by .0, like 0.0 and 1.0.
TreeClassification
|
Parameter |
Type & Range |
Description |
|
criterion |
string: ["gini", "entropy"] |
Criterions for evaluating node splitting. |
|
splitter |
string: ["best", "random"] |
Choose a splitting strategy for each node. |
|
max_depth |
int: [1, +∞), {1, 100, 1} null |
Maximum tree depth. |
|
min_samples_split |
int: [1, +∞), {10, 1000, 10} float: (0, 1) |
Minimum amount sampling data for node splitting. int is the min amount of sampling data and float is the proportion of it to the whole data. |
|
min_samples_leaf |
int: [1, +∞), {10, 1000, 10} float: (0, 1) |
Minimum amount of sampling data for leaf node. int is the min amount of sampling data and float is the proportion of it to the whole data. |
|
min_weight_fraction_leaf |
float: [0, 1), {0, 0.1, 0.01} |
Minimum weight among all weights of the input sampling data at a leaf node. |
|
max_features |
int: [1, +∞), {10, 1000, 10} float: (0, 1] string: ["auto", "sqrt", "log2"] null |
Get the maximum number of variables for the optimal node splitting. Max number of variables for an int parameter. Max proportion of variables for a float parameter. If "auto", then max_features=sqrt(n_features). If "sqrt", then max_features=sqrt(n_features). If "log2", then max_features=log2(n_features). If null, then max_features=n_features. |
|
max_leaf_nodes |
int: [1, +∞), {10, 1000, 10} null |
Use best-first fashion to generate the largest number of leaf nodes in a pruned tree. null means that there’s no limitation on the number of leaf nodes. |
|
min_impurity_decrease |
float: [0, 1) |
The lowest impurity decrease for node splitting. |
|
class_weight |
string: [“balanced”] null |
Weights associated with classes in the form {class_label: weight}. |
GBDTClassification
|
Parameter |
Type |
Description |
|
loss |
string: ["deviance", "exponential"] |
A loss function. |
|
learning_rate |
float: (0, 1), {0.1, 0.9, 0.1} |
The learning rate, which is in direct ratio to the training speed. But it’s probably that there isn’t an optimal solution. |
|
n_estimators |
int: [1, +∞), {10, 500, 10} |
Number of boosting stages. |
|
subsample |
float: (0, 1], {0.1, 1, 0.1} |
The ratio of sampling data used by a basic machine learning method. |
|
criterion |
string: ["mse", "friedman_mse", "mae"] |
Criterions for evaluating node splitting. |
|
min_samples_split |
int: [1, +∞), {10, 1000, 10} float: (0, 1) |
Minimum amount sampling data for node splitting. int is the min amount of sampling data and float is the proportion of it to the whole data. |
|
min_samples_leaf |
int: [1, +∞), {10, 1000, 10} float: (0, 1) |
Minimum amount of sampling data for leaf node. int is the min amount of sampling data and float is the proportion of it to the whole data. |
|
min_weight_fraction_leaf |
float: [0, 1), {0, 0.1, 0.01} |
Minimum weight among all weights of the input sampling data at a leaf node. |
|
max_depth |
int: [1, +∞), {1, 100, 1} null |
Maximum tree depth. |
|
min_impurity_decrease |
float: [0, 1) |
The lowest impurity decrease for node splitting. |
|
max_features |
int: [1, +∞), {10, 1000, 10} float: (0, 1] string: ["auto", "sqrt", "log2"] null |
Get the maximum number of variables for the optimal node splitting. Max number of variables for an int parameter. Max proportion of variables for a float parameter. If "auto", then max_features=sqrt(n_features). If "sqrt", then max_features=sqrt(n_features). If "log2", then max_features=log2(n_features). If null, then max_features=n_features. |
|
max_leaf_nodes |
int: [1, +∞), {10, 1000, 10} null |
Use best-first fashion to generate the largest number of leaf nodes in a pruned tree. null means that there’s no limitation on the number of leaf nodes. |
|
warm_start |
bool |
Use the result of the previous iteration if the value is true; and won’t use that if the value is false. |
RFClassification
|
Parameter |
Type |
Description |
|
n_estimators |
int: [1, +∞), {10, 500, 10} |
The number of trees. |
|
criterion |
string: ["gini", "entropy"] |
Criterions for evaluating node splitting. |
|
max_depth |
int: [1, +∞), {1, 100, 1} null |
Maximum tree depth. |
|
min_samples_split |
int: [1, +∞), {10, 1000, 10} float: (0, 1) |
Minimum amount sampling data for node splitting. int is the min amount of sampling data and float is the proportion of it to the whole data. |
|
min_samples_leaf |
int: [1, +∞), {10, 1000, 10} float: (0, 1) |
Minimum amount of sampling data for leaf node. int is the min amount of sampling data and float is the proportion of it to the whole data. |
|
min_weight_fraction_leaf |
float: [0, 1), {0, 0.1, 0.01} |
Minimum weight among all weights of the input sampling data at a leaf node. |
|
max_features |
int: [1, +∞), {10, 1000, 10} float: (0, 1] string: ["auto", "sqrt", "log2"] null |
Get the maximum number of variables for the optimal node splitting. Max number of variables for an int parameter. Max proportion of variables for a float parameter. If "auto", then max_features=sqrt(n_features). If "sqrt", then max_features=sqrt(n_features). If "log2", then max_features=log2(n_features). If null, then max_features=n_features. |
|
max_leaf_nodes |
int: [1, +∞), {10, 1000, 10} null |
Use best-first fashion to generate the largest number of leaf nodes in a pruned tree. null means that there’s no limitation on the number of leaf nodes. |
|
min_impurity_decrease |
float: [0, 1) |
The lowest impurity decrease for node splitting. |
|
bootstrap |
bool |
Whether to use bootstrap when generating a tree. |
|
oob_score |
bool |
Whether to use out-of-bag samples to predict accuracy. |
|
warm_start |
bool |
Use the result of the previous iteration if the value is true; and won’t use that if the value is false. |
|
class_weight |
string: [“balanced”] null |
Weights associated with classes in the form {class_label: weight}. |
LogicClassification
|
Parameter |
Type |
Description |
|
penalty |
string: ["l1", "l2", "elasticnet", "none"] |
Penalty regularization. "newton-cg", "sag" and "lbfgs" solvers support "l2" only; "elasticnet" supports ‘saga’ solver only; "none" means non-regularization and doesn’t support liblinear solver. |
|
dual |
bool |
Dual or primal formulation. Dual formulation is only implemented for l2 penalty with liblinear solver. Prefer dual=False when n_samples > n_features. |
|
tol |
float: (0, 1) |
The tolerance value before stopping iteration. |
|
C |
float: (0, 1] |
Inverse of regularization strength, which must be a positive. |
|
fit_intercept |
bool |
Whether to include an intercept item. |
|
intercept_scaling |
float: (0, 1] |
Only works when solver="liblinear". |
|
class_weight |
string: [“balanced”] null |
Weights associated with classes in the form {class_label: weight}. |
|
solver |
string: ["newton-cg", "lbfgs", "liblinear", "sag", "saga"] |
The optimal algorithm. |
|
max_iter |
int: [1, +∞), {10, 500, 10} |
Maximum iterations; only works when solver=["newton-cg", "lbfgs", "sag"]. |
|
multi_class |
string: ["ovr", "multinomial", "auto"] |
The algorithm for handling multiple classes. "ovr" builds a model for each class; "multinomial" doesn’t work with solver="liblinear". |
|
warm_start |
bool |
Use the result of the previous iteration if the value is true; and won’t use that if the value is false. |
RidgeClassification
|
Parameter |
Type |
Description |
|
alpha |
float:[0, +∞], {0.0, 10.0, 0.1} |
Regularization strength, which must be a positive. |
|
fit_intercept |
bool |
Whether to include an intercept item. |
|
normalize |
bool |
Whether to normalize data. |
|
max_iter |
int: [1, +∞), {10, 500, 10} null |
Maximum iterations. |
|
tol |
float: (0, 1) |
Precision of the final solution. |
|
class_weight |
string: [“balanced”] null |
Weights associated with classes in the form {class_label: weight}. If not given, all classes are supposed to have weight one. "balanced" for auto-adjust. |
|
solver |
string: ["auto", "svd", "cholesky", "lsqr", "sparse_cg", "sag", "saga"] |
The optimal algorithm. |
FNNClassification and CNNClassification
Parameters for these two types of binary model are not supported for the time being due to some special features of the neural networks.
XGBClassification
|
Parameter |
Type |
Description |
|
max_depth |
int: [1, +∞), {1, 100, 1} |
Maximum tree depth. |
|
learning_rate |
float: (0, 1), {0.1, 0.9, 0.1} |
The learning rate, which is in direct ratio to the training speed. But it’s probably that there isn’t an optimal solution. |
|
n_estimators |
int: [1, +∞), {10, 500, 10} |
Number of booster trees. |
|
objective |
string: ["binary:logistic", "binary:logitraw", "binary:hinge"] |
Learning objective; binary:logistic: Binary logistic regression for outputting probability; binary:logitraw: Binary logistic regression for outputting the score before logistic transformation; binary:hinge: Binary hinge loss for outputting class 0 or class 1 instead of the probability. |
|
booster |
string: ["gbtree", "gblinear", "dart"] |
The booster type used. |
|
gamma |
float: [0, +∞) |
The smallest loss mitigation value for node splitting. |
|
min_child_weight |
int: [1, +∞), {10, 1000, 10} |
The minimum sum of sampling weights of child nodes. |
|
max_delta_step |
int: [0, +∞), {0, 10, 1} |
The allowed longest delta step for evaluating a tree’s weight. |
|
subsample |
float: (0, 1], {0.1, 1.0, 0.1} |
The proportion of subsample for training a model to the whole set of samplings. |
|
colsample_bytree |
float: (0, 1], {0.1, 1.0, 0.1} |
Proportion of the random sampling from the features for each tree. |
|
colsample_bylevel |
float: (0, 1], {0.1, 1.0, 0.1} |
Proportion of random sampling from the features on each horizontal level for node splitting. |
|
reg_alpha |
float:[0, +∞], {0.0, 10.0, 0.1} |
L1 regularization term. |
|
reg_lambda |
float:[0, +∞], {0.0, 10.0, 0.1} |
L2 regularization term. |
|
scale_pos_weight |
float: (0, +∞) |
Control the balance of positive samples and negative samples. |
|
base_score |
float: (0, 1), {0.1, 0.9, 0.1} |
The initial value for starting a prediction. |
|
missing |
float: (-∞, +∞) null |
Define a missing value. |
PCAClassification
|
Parameter |
Type |
Description |
|
n_components |
int or null: [1, min(row count, column count )] |
Retain the number of principal components; null indicates auto-config, which is the default. |
|
whiten |
bool |
Whether to convert unit root. |
|
svd_solver |
string: ["auto", "full", "arpack", "randomized"] |
The SVD solver to find PCA; default is full. |
|
tol |
float: (0, 1) |
Tolerance to use; default is 0.0001. |
|
fit_intercept |
bool |
Whether to include an intercept item. |
|
max_iter |
int: [1, +∞), {100, 1000, 100} |
Maximum number of iterations. |
|
reg_solver |
string: ["newton-cg", "lbfgs", "sag", "saga"] |
The regression solver to find PCA; default is "lbfgs". |
|
warm_start |
bool |
Use the result of the previous iteration if the value is true; and won’t use that if the value is false. |
Regression model
You can configure parameters for regression models on “Regression model” tab. A selected regression model will be used for model building.
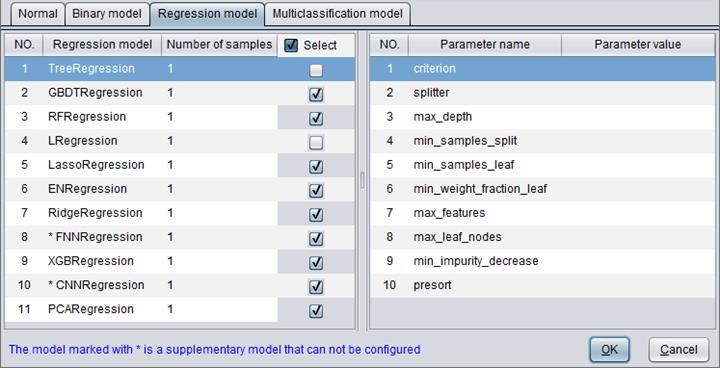
There are 11 types of regression models – TreeRegression, GBDTRegression, RFRegression, LRegression, LassoRegression, ENRegression, RidgeRegression, FNNRegression, XGBRegression, CNNRegression and PCARegression.
“Number of samples” determines the number of samples used to build a model.
Below are parameter configuration directions for regression models.
Appendix 2: Regression model parameters
Type & Range: The type is always followed by an interval indicating the parameter’s value range. Square brackets indicate a closed interval and parentheses indicate an opened interval. Braces are used for certain int and float parameters to represent drop-down-menu values; the format is {start value, end value, interval}, such as {1, 5, 1}=[1,2,3,4,5] and {1, 6, 2}=[1,3,5]. All available values of string parameters will be listed in the drop-down menu and can’t be entered manually; a null value should be selected through the drop-down menu; the bool values, which are true and false, also should be selected in the drop-down menu; values of int parameters and float parameters can be enterned manually and all available values will be listed in drop-down menu.
For some float parameters, if their values are integers, they need to be followed by .0, like 0.0 and 1.0.
TreeRegression
|
Parameter |
Type |
Description |
|
criterion |
string: ["mse", "friedman_mse", "mae"] |
Criterions for evaluating node splitting. |
|
splitter |
string: ["best", "random"] |
Choose a splitting strategy for each node. |
|
max_depth |
int: [1, +∞), {1, 100, 1} null |
Maximum tree depth. |
|
min_samples_split |
int: [1, +∞), {10, 1000, 10} float: (0, 1) |
Minimum amount sampling data for node splitting. int is the min amount of sampling data and float is the proportion of it to the whole data. |
|
min_samples_leaf |
int: [1, +∞), {10, 1000, 10} float: (0, 1) |
Minimum amount of sampling data for leaf node. int is the min amount of sampling data and float is the proportion of it to the whole data. |
|
min_weight_fraction_leaf |
float: [0, 1), {0, 0.1, 0.01} |
Minimum weight among all weights of the input sampling data at a leaf node. |
|
max_features |
int: [1, +∞), {10, 1000, 10} float: (0, 1] string: ["auto", "sqrt", "log2"] null |
Get the maximum number of variables for the optimal node splitting. Max number of variables for an int parameter. Max proportion of variables for a float parameter. If "auto", then max_features=sqrt(n_features). If "sqrt", then max_features=sqrt(n_features). If "log2", then max_features=log2(n_features). If null, then max_features=n_features. |
|
max_leaf_nodes |
int: [1, +∞), {10, 1000, 10} null |
Use best-first fashion to generate the largest number of leaf nodes in a pruned tree. null means that there’s no limitation on the number of leaf nodes. |
|
min_impurity_decrease |
float: [0, 1) |
The lowest impurity decrease for node splitting. |
GBDTRegression
|
Parameter |
Type |
Description |
|
loss |
string: ["ls", "lad", "huber", "quantile"] |
A loss function. |
|
learning_rate |
float: (0, 1), {0.1, 0.9, 0.1} |
The learning rate, which is in direct ratio to the training speed. But it’s probably that there isn’t an optimal solution. |
|
n_estimators |
int: [1, +∞), {10, 500, 10} |
Number of boosting stages. |
|
subsample |
float: (0, 1], {0.1, 1, 0.1} |
The ratio of sampling data used by a basic machine learning method. |
|
criterion |
string: ["mse", "friedman_mse", "mae"] |
Criterions for evaluating node splitting. |
|
min_samples_split |
int: [1, +∞), {10, 1000, 10} float: (0, 1) |
Minimum amount sampling data for node splitting. int is the min amount of sampling data and float is the proportion of it to the whole data. |
|
min_samples_leaf |
int: [1, +∞), {10, 1000, 10} float: (0, 1) |
Minimum amount of sampling data for leaf node. int is the min amount of sampling data and float is the proportion of it to the whole data. |
|
min_weight_fraction_leaf |
float: [0, 1), {0, 0.1, 0.01} |
Minimum weight among all weights of the input sampling data at a leaf node. |
|
max_depth |
int: [1, +∞), {1, 100, 1} null |
Maximum tree depth. |
|
min_impurity_decrease |
float: [0, 1) |
The lowest impurity decrease for node splitting. |
|
max_features |
int: [1, +∞), {10, 1000, 10} float: (0, 1] string: ["auto", "sqrt", "log2"] null |
Get the maximum number of variables for the optimal node splitting. Max number of variables for an int parameter. Max proportion of variables for a float parameter. If "auto", then max_features=sqrt(n_features). If "sqrt", then max_features=sqrt(n_features). If "log2", then max_features=log2(n_features). If null, then max_features=n_features. |
|
alpha |
float: (0, 1), {0.1, 0.9, 0.1} |
The alpha-quantile of the huber loss function and the quantile loss function. Only if loss='huber' or loss='quantile' |
|
max_leaf_nodes |
int: [1, +∞), {10, 1000, 10} null |
Use best-first fashion to generate the largest number of leaf nodes in a pruned tree. null means that there’s no limitation on the number of leaf nodes. |
|
warm_start |
bool |
Use the result of the previous iteration if the value is true; and won’t use that if the value is false. |
RFRegression
|
Parameter |
Type |
Description |
|
n_estimators |
int: [1, +∞), {10, 500, 10} |
The number of trees. |
|
criterion |
string: ["mse", "mae"] |
Criterions for evaluating node splitting. |
|
max_depth |
int: [1, +∞), {1, 100, 1} null |
Maximum tree depth. |
|
min_samples_split |
int: [1, +∞), {10, 1000, 10} float: (0, 1) |
Minimum amount sampling data for node splitting. int is the min amount of sampling data and float is the proportion of it to the whole data. |
|
min_samples_leaf |
int: [1, +∞), {10, 1000, 10} float: (0, 1) |
Minimum amount of sampling data for leaf node. int is the min amount of sampling data and float is the proportion of it to the whole data. |
|
min_weight_fraction_leaf |
float: [0, 1), {0, 0.1, 0.01} |
Minimum weight among all weights of the input sampling data at a leaf node. |
|
max_features |
int: [1, +∞), {10, 1000, 10} float: (0, 1] string: ["auto", "sqrt", "log2"] null |
Get the maximum number of variables for the optimal node splitting. Max number of variables for an int parameter. Max proportion of variables for a float parameter. If "auto", then max_features=sqrt(n_features). If "sqrt", then max_features=sqrt(n_features). If "log2", then max_features=log2(n_features). If null, then max_features=n_features. |
|
max_leaf_nodes |
int: [1, +∞), {10, 1000, 10} null |
Use best-first fashion to generate the largest number of leaf nodes in a pruned tree. null means that there’s no limitation on the number of leaf nodes. |
|
min_impurity_decrease |
float: [0, 1) |
The lowest impurity decrease for node splitting. |
|
bootstrap |
bool |
Whether to use bootstrap when generating a tree. |
|
oob_score |
bool |
Whether to use out-of-bag samples to predict accuracy. |
|
warm_start |
bool |
Use the result of the previous iteration if the value is true; and won’t use that if the value is false. |
LRegression
|
Parameter |
Type |
Description |
|
fit_intercept |
bool |
Whether to include an intercept item. |
|
normalize |
bool |
Whether to normalize data. |
LassoRegression
|
Parameter |
Type |
Description |
|
fit_intercept |
bool |
Whether to include an intercept item. |
|
alpha |
float or null:[0, +∞], {0.0, 10.0, 0.1} |
The regularized penalty factor. A null value means auto-configure and a float will disable cv and max_n_alphas. |
|
normalize |
bool |
Whether to normalize data. |
|
precompute |
string: ["auto"] bool |
Whether to precompute Gram matrix to speed up model building. |
|
max_iter |
int: [1, +∞), {10, 500, 10} |
Maximum number of iterations. |
|
cv |
int: [2, 20] |
Cross-validate the turning point |
|
max_n_alphas |
int: [1, +∞) , {100, 1000, 100} |
Cross-validate the number of searched alpha |
|
positive |
bool |
Whether to rule that a coefficient must be positive. |
ENRegression
|
Parameter |
Type |
Description |
|
alpha |
float or null: [0, +∞], {0.0, 10.0, 0.1} |
A constant multiplied by penalty item; null indicates auto-config, which is the default. |
|
l1_ratio |
float or null : [0, 1], {0.0, 1.0, 0.1} |
A mixed parameter; its value is L2 when l1_ratio=0; and its value is L1 when l1_ratio=1; its value is mixed proportion when 11_ratio falls between 0 and 1; null indicates auto-config, which is the default. |
|
n_alphas |
int: [1, +∞), {100, 1000, 100} |
Get the number of alpha; it is invalid when alpha is a float. |
|
cv |
int: [2, 20] |
Cross-validate the turning point; it is invalid when both alpha and l1_ratio are float. |
|
fit_intercept |
bool |
Whether to include an intercept item. |
|
normalize |
bool |
Whether to normalize data. |
|
precompute |
bool |
Whether to precompute Gram matrix to speed up model building. |
|
max_iter |
int: [1, +∞), {10, 500, 10} |
Maximum iterations. |
|
tol |
float: (0, 1) |
The tolerance value before stopping iteration. |
|
warm_start |
bool |
Use the result of the previous iteration if the value is true; and won’t use that if the value is false. It works when parameter cv is disabled; and it is invalid when cv works. |
|
positive |
bool |
Whether to rule that a coefficient must be positive. |
|
selection |
string: ["cyclic", "random"] |
"cyclic" means iteration by loop by variables; "random" represents a random iteration coefficient. |
RidgeRegression
|
Parameter |
Type |
Description |
|
alpha |
float: [0, +∞], {0.0, 10.0, 0.1} |
Regularization strength, which must be a positive. |
|
fit_intercept |
bool |
Whether to include an intercept item. |
|
normalize |
bool |
Whether to normalize data. |
|
max_iter |
int: [1, +∞), {10, 500, 10} null |
Maximum iterations. |
|
tol |
float: (0, 1) |
Precision of the final solution. |
|
solver |
string: ["auto", "svd", "cholesky", "lsqr", "sparse_cg", "sag", "saga"] |
The optimal algorithm. |
FNNRegression and CNNRegression
Parameters for these two types of regression model are not supported for the time being due to some special features of the neural networks.
XGBRegression
|
Parameter |
Type |
Description |
|
max_depth |
int: [1, +∞), {1, 100, 1} |
Maximum tree depth. |
|
learning_rate |
float: (0, 1), {0.1, 0.9, 0.1} |
The learning rate, which is in direct ratio to the training speed. But it’s probably that there isn’t an optimal solution. |
|
n_estimators |
int: [1, +∞), {10, 500, 10} |
Number of booster trees. |
|
objective |
string: ["reg:squarederror", "reg:squaredlogerror", "reg:logistic"] |
Learning objective; reg:squarederror: Squared error loss; reg:squaredlogerror: Squared log error loss; reg:logistic: logistic regression. |
|
booster |
string: ["gbtree", "gblinear", "dart"] |
The booster type used. |
|
gamma |
float: [0, +∞) |
The smallest loss mitigation value for node splitting. |
|
min_child_weight |
int: [1, +∞), {10, 1000, 10} |
The minimum sum of sampling weights of child nodes. |
|
max_delta_step |
int: [0, +∞), {0, 10, 1} |
The allowed longest delta step for evaluating a tree’s weight. |
|
subsample |
float: (0, 1], {0.1, 1.0, 0.1} |
The proportion of subsample for training a model to the whole set of samplings. |
|
colsample_bytree |
float: (0, 1], {0.1, 1.0, 0.1} |
Proportion of the random sampling from the features for each tree. |
|
colsample_bylevel |
float: (0, 1], {0.1, 1.0, 0.1} |
Proportion of random sampling from the features on each horizontal level for node splitting. |
|
reg_alpha |
float:[0, +∞], {0.0, 10.0, 0.1} |
L1 regularization term. |
|
reg_lambda |
float:[0, +∞], {0.0, 10.0, 0.1} |
L2 regularization term. |
|
scale_pos_weight |
float: (0, +∞) |
Control the balance of positive samples and negative samples. |
|
base_score |
float: (0, 1), {0.1, 0.9, 0.1} |
The initial value for starting a prediction. |
|
missing |
float: (-∞, +∞) null |
Define a missing value. |
PCARegression
|
Parameter |
Type |
Description |
|
n_components |
int or null: [1, min(row count, column count)] |
Retain the number of principal components; null indicates auto-config, which is the default. |
|
whiten |
bool |
Whether to convert unit root. |
|
svd_solver |
string: ["auto", "full", "arpack", "randomized"] |
The SVD solver to find PCA; default is full. |
|
tol |
float: (0, 1) |
Tolerance to use; default is 0.0001. |
|
fit_intercept |
bool |
Whether to include an intercept item. |
|
normalize |
bool |
Whether to normalize data. |
Multi-category model
“Multi-category model”: It is for configuring the multi-category model. A selected multi-category model will be used for model building.
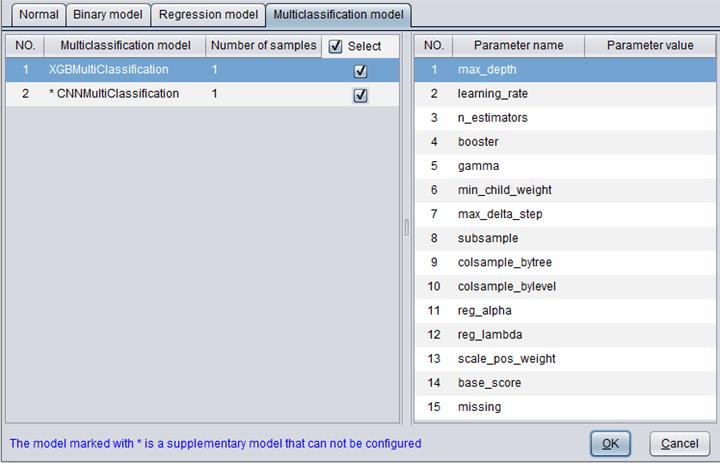
There are two types of multi-category model – XGBMultiClassification、CNNMultiClassification.
The “Number of samples” specifies the count of samples to score data according to a certain model.
The following Appendix 3 lists parameters and their descriptions for the multi-category model.
Appendix 3: multi-category model parameters
XGBMultiClassification
|
Parameter |
Type |
Description |
|
max_depth |
int: [1, +∞), {1, 100, 1} |
Maximum tree depth |
|
learning_rate |
float: (0, 1), {0.1, 0.9, 0.1} |
The learning rate, which is in direct ratio to the training speed. But it’s probably that there isn’t an optimal solution. |
|
n_estimators |
int: [1, +∞), {10, 500, 10} |
The number of trees. |
|
booster |
string: ["gbtree", "gblinear", "dart"] |
The booster type used. |
|
gamma |
float: [0, +∞) |
The smallest loss mitigation value for node splitting. |
|
min_child_weight |
int: [1, +∞), {10, 1000, 10} |
The minimum sum of sampling weights of child nodes. |
|
max_delta_step |
int: [0, +∞), {0, 10, 1} |
The allowed longest delta step for evaluating a tree’s weight. |
|
subsample |
float: (0, 1], {0.1, 1.0, 0.1} |
The proportion of subsample for training a model to the whole set of samplings. |
|
colsample_bytree |
float: (0, 1], {0.1, 1.0, 0.1} |
Proportion of the random sampling from the features for each tree. |
|
colsample_bylevel |
float: (0, 1], {0.1, 1.0, 0.1} |
Proportion of random sampling from the features on each horizontal level for node splitting. |
|
reg_alpha |
float:[0, +∞], {0.0, 10.0, 0.1} |
L1 regularization term. |
|
reg_lambda |
float:[0, +∞], {0.0, 10.0, 0.1} |
L2 regularization term. |
|
scale_pos_weight |
float: (0, +∞) |
Control the balance of positive samples and negative samples. |
|
base_score |
float: (0, 1), {0.1, 0.9, 0.1} |
The initial value for starting a prediction. |
|
missing |
float: (-∞, +∞) null |
Define a missing value. |
CNNMultiClassification
This type of parameters is not supported for the time being due to some special features of the neural networks.
Execute model building
To build a predictive model, you must choose a target variable and then select a modeling table file through “Model file”. By default the modeling table file is stored under the same directory where the loaded data is stored and has the same name as the loaded data file. Users can define the path and name themselves. A model document is one with .pcf extension.
![]()
Click “Modeling” option
under “Run”, or click ![]() on the toolbar, to pop up the “Build model” window, where model
building information is output.
on the toolbar, to pop up the “Build model” window, where model
building information is output.
Model building is finished when the message “Model building is finished” appears.
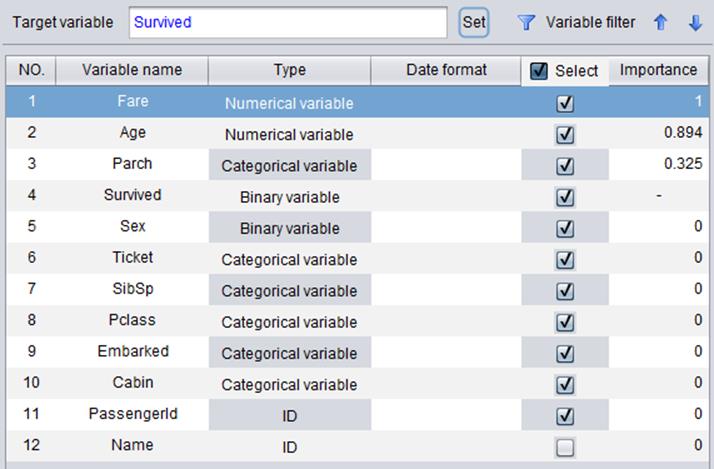
“Importance” will be displayed after a model is built, as shown below. A variable’s degree of importance indicates its influence on the future predictive result. The higher the degree of importance is, the bigger a variable’s influence is. A variable with zero importance degree has no impact on the predictive result. As the following shows, the Sex variable has the biggest influence on the result.
Model file information
Model presentation
YModel encapsulates multiple algorithms for model building, they are: Decision Tree, Gradient Boosting, Logistic Regression, Neural Network, Random Forest, Elastic Net, LASSO Regression, Linear Regression, Ridge Regression, and XGBoost.
After the model building is finished, you can click “Model presentation” in “Build model” window to view the algorithm(s) used to build the model, as shown below:
Model performance
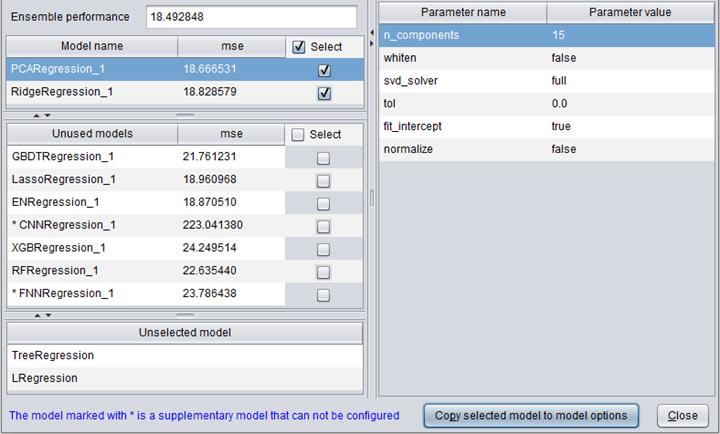
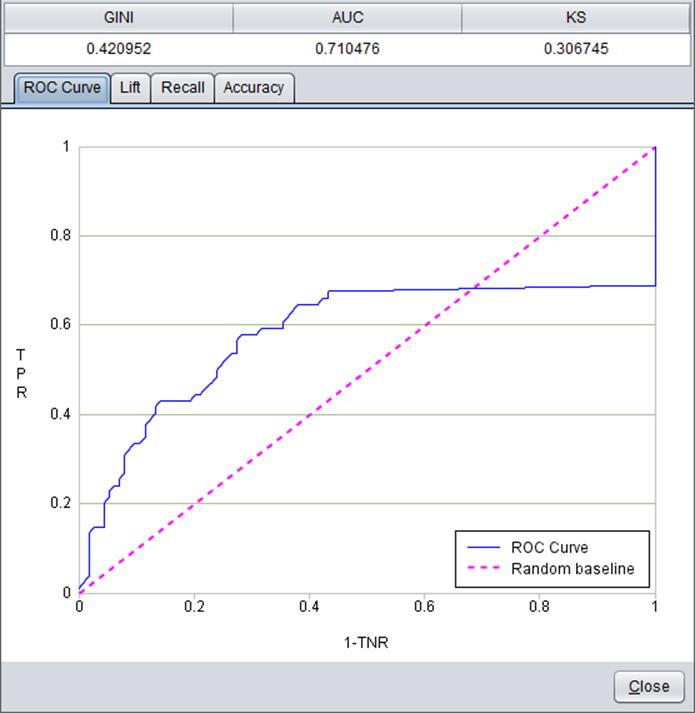
Model performance can be reflected through a series of indexes and figures.
Click “Model performance” in “Build model” window:
![]()
Models built on different types of target variables are evaluated by different parameters and forms.
Below is model performance information about binary target variable “Survived”:
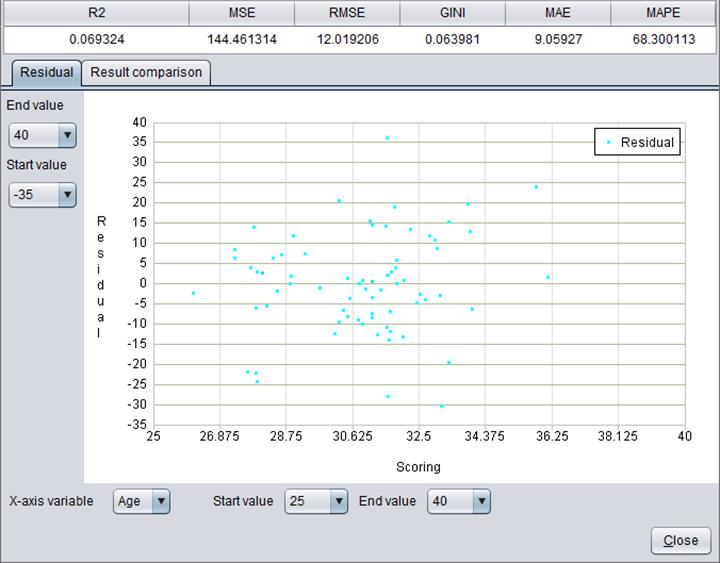
Here’s model performance information about numerical target variable “Age”: